041 GDAL: time series : Answers to exercises
Exercise 1
We have seen in 040_GDAL_mosaicing_and_masking that you can use gdal to creat a GeoTiff format image, for example with:
g = gdal.Warp(output_name, input_name ,format='GTiff',options=['COMPRESS=LZW'])
g.FlushCache()
- Convert the
gdalfilework/stitch_set.vrtto a more portable GeoTiff file calledwork/stitch_set.tif - Confirm that this has worked by reading and displaying data from the file
# ANSWER
from osgeo import gdal
# Convert the `gdal` file `work/stitch_set.vrt` to a
# more portable GeoTiff file called `work/stitch_set.tif`
# set up the filenames
infile = 'work/stitch_set.vrt'
outfile = 'work/stitch_set.tif'
# convert using gdal.Warp or similar
g = gdal.Warp(outfile, infile ,format='GTiff',options=['COMPRESS=LZW'])
# force write to disk
g.FlushCache()
from osgeo import gdal
import matplotlib.pyplot as plt
# ANSWER
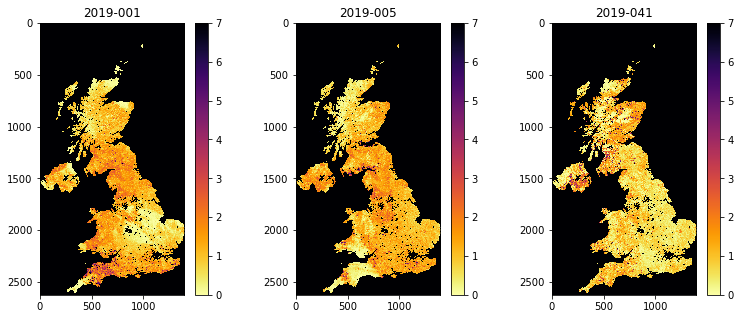
fig, axs = plt.subplots(1,3,figsize=(13,5))
axs = axs.flatten()
for i in range(data.shape[0]):
im = axs[i].imshow(data[i],vmax=7,\
cmap=plt.cm.inferno_r,interpolation='nearest')
fig.colorbar(im, ax=axs[i])
axs[i].set_title(bnames[i])
Exercise 2
- Produce a set of spatial plots of the quantity
Fpar_500mover Luxembourg for 2019
# ANSWER
msg = '''
Produce a set of spatial
plots of the quantity `Fpar_500m` over Luxembourg for 2019
This is almost identical to the above, but with a different SDS
'''
from geog0111.modisUtils import getModis
from osgeo import gdal
warp_args = {
'dstNodata' : 255,
'format' : 'MEM',
'cropToCutline' : True,
'cutlineWhere' : "FIPS='LU'",
'cutlineDSName' : 'data/TM_WORLD_BORDERS-0.3.shp'
}
kwargs = {
'tile' : ['h18v03','h18v04'],
'product' : 'MCD15A3H',
'sds' : 'Fpar_500m',
'doys' : np.arange(1,366,4),
'year' : 2019,
'warp_args' : warp_args
}
datafiles,bnames = getModis(verbose=False,timeout=None,**kwargs)
# build VRT
stitch_vrt = gdal.BuildVRT("work/stitch_time.vrt", datafiles,separate=True)
del stitch_vrt
# read data
g = gdal.Warp("","work/stitch_time.vrt",**warp_args)
data = g.ReadAsArray() * 0.1
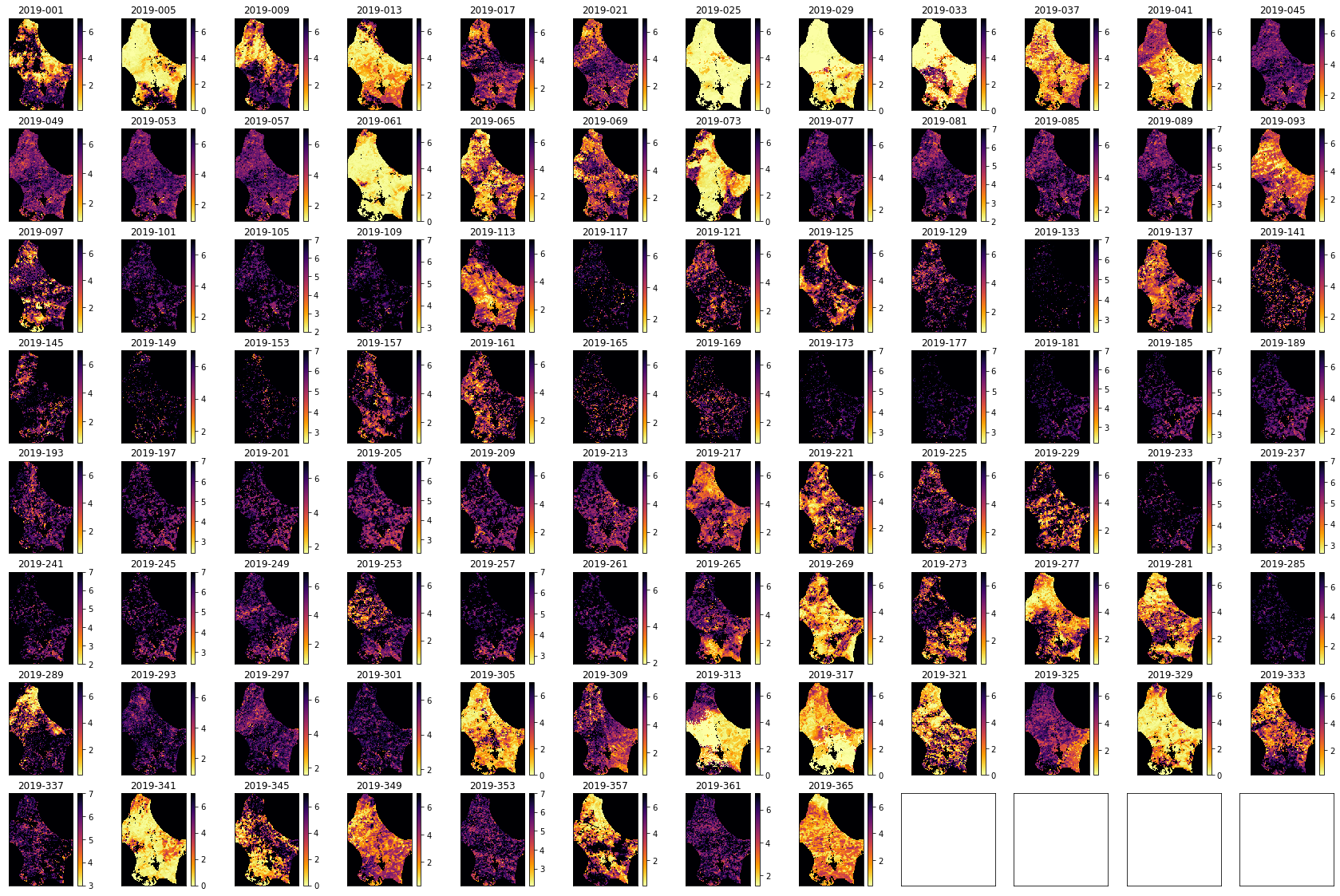
import matplotlib.pyplot as plt
shape=(8,12)
x_size,y_size=(30,20)
fig, axs = plt.subplots(*shape,figsize=(x_size,y_size))
axs = axs.flatten()
plt.setp(axs, xticks=[], yticks=[])
for i in range(data.shape[0]):
im = axs[i].imshow(data[i],vmax=7,cmap=plt.cm.inferno_r,\
interpolation='nearest')
axs[i].set_title(bnames[i])
fig.colorbar(im, ax=axs[i])
Exercise 3
Write a function called modisAnnual(**kwargs) with arguments based on:
warp_args = {
'dstNodata' : 255,
'format' : 'MEM',
'cropToCutline' : True,
'cutlineWhere' : "FIPS='LU'",
'cutlineDSName' : 'data/TM_WORLD_BORDERS-0.3.shp'
}
kwargs = {
'tile' : ['h18v03','h18v04'],
'product' : 'MCD15A3H',
'sds' : ['Lai_500m', 'Fpar_500m'],
'doys' : np.arange(1,366,4),
'year' : 2019,
'warp_args' : warp_args
'ofile_root': 'work/output_filename_ex3'
}
where sds is a list of SDS
That returns:
bnames : names for the items in first (time) dimension
odict : a dictionary with items in sds for keys and the names of associated VRT files as values
from geog0111.modisUtils import getModis
from pathlib import Path
from osgeo import gdal
#ANSWER
msg = '''This is almost the same as the previous exercise, but wrapped as a function with a loop around SDS'''
def modisAnnual(ofile_root='work/output_filename',**kwargs):
'''
generate dictionary of SDS datasets as VRT files
arguments based on:
warp_args = {
'dstNodata' : 255,
'format' : 'MEM',
'cropToCutline' : True,
'cutlineWhere' : "FIPS='LU'",
'cutlineDSName' : 'data/TM_WORLD_BORDERS-0.3.shp'
}
kwargs = {
'tile' : ['h18v03','h18v04'],
'product' : 'MCD15A3H',
'sds' : ['Lai_500m', 'Fpar_500m'],
'doys' : np.arange(1,366,4),
'year' : 2019,
'warp_args' : warp_args
}
Return
'''
sds_list = kwargs['sds']
# output dict
odict = {}
for s in sds_list:
ofile = f"{ofile_root}_{s}.vrt"
kwargs['sds'] = s
datafiles,bnames = getModis(**kwargs)
stitch_vrt = gdal.BuildVRT(ofile, datafiles,separate=True)
# save the band names
bofile = Path(f'{ofile}_bands')
bofile.write_text(' '.join(bnames))
del stitch_vrt
odict[s] = ofile
return odict,bnames
print(msg)
This is almost the same as the previous exercise, but wrapped as a function with a loop around SDS
#ANSWER
#slightly better as does checking to see if file exists
# and we store the bnames data
def modisAnnual(ofile_root='work/output_filename',**kwargs):
'''
generate dictionary of SDS datasets as VRT files
arguments based on:
warp_args = {
'dstNodata' : 255,
'format' : 'MEM',
'cropToCutline' : True,
'cutlineWhere' : "FIPS='LU'",
'cutlineDSName' : 'data/TM_WORLD_BORDERS-0.3.shp'
}
kwargs = {
'tile' : ['h18v03','h18v04'],
'product' : 'MCD15A3H',
'sds' : ['Lai_500m', 'Fpar_500m'],
'doys' : np.arange(1,60,4),
'year' : 2019,
'warp_args' : warp_args
}
Return odict,bnames
where odict keys are SDS values and the values VRT filenames
'''
sds_list = kwargs['sds']
# output dict
odict = {}
if ('force' in kwargs.keys()) and kwargs['force'] == True:
redo = True
del kwargs['force']
else:
redo = False
bnames = []
for s in sds_list:
ofile = f"{ofile_root}_SDS_{s}.vrt"
bofile = Path(f'{ofile}_bands')
if not redo:
if (not Path(ofile).exists()) or (not bofile.exists()):
kwargs['sds'] = s
datafiles,bnames = getModis(**kwargs)
stitch_vrt = gdal.BuildVRT(ofile, datafiles,separate=True)
# save the band names
bofile = Path(f'{ofile}_bands')
bofile.write_text(' '.join(bnames))
del stitch_vrt
else:
kwargs['sds'] = s
datafiles,bnames = getModis(**kwargs)
stitch_vrt = gdal.BuildVRT(ofile, datafiles,separate=True)
del stitch_vrt
# save the band names
bofile = Path(f'{ofile}_bands')
bofile.write_text(' '.join(bnames))
odict[s] = ofile
bofile = Path(f'{ofile}_bands')
bnames = bofile.read_text().split()
return odict,bnames
#ANSWER
# test
warp_args = {
'dstNodata' : 255,
'format' : 'MEM',
'cropToCutline' : True,
'cutlineWhere' : "FIPS='LU'",
'cutlineDSName' : 'data/TM_WORLD_BORDERS-0.3.shp'
}
kwargs = {
'tile' : ['h18v03','h18v04'],
'product' : 'MCD15A3H',
'sds' : ['Lai_500m', 'Fpar_500m'],
'doys' : np.arange(1,366,4),
'year' : 2019,
'verbose' : False,
'timeout' : None,
'ofile_root': 'work/output_filename_ex3',
'warp_args' : warp_args
}
# run
odict,bnames = modisAnnual(**kwargs)
# outputs
print(odict,bnames)
{'Lai_500m': 'work/output_filename_ex3_SDS_Lai_500m.vrt', 'Fpar_500m': 'work/output_filename_ex3_SDS_Fpar_500m.vrt'} ['2019-001', '2019-005', '2019-009', '2019-013', '2019-017', '2019-021', '2019-025', '2019-029', '2019-033', '2019-037', '2019-041', '2019-045', '2019-049', '2019-053', '2019-057', '2019-061', '2019-065', '2019-069', '2019-073', '2019-077', '2019-081', '2019-085', '2019-089', '2019-093', '2019-097', '2019-101', '2019-105', '2019-109', '2019-113', '2019-117', '2019-121', '2019-125', '2019-129', '2019-133', '2019-137', '2019-141', '2019-145', '2019-149', '2019-153', '2019-157', '2019-161', '2019-165', '2019-169', '2019-173', '2019-177', '2019-181', '2019-185', '2019-189', '2019-193', '2019-197', '2019-201', '2019-205', '2019-209', '2019-213', '2019-217', '2019-221', '2019-225', '2019-229', '2019-233', '2019-237', '2019-241', '2019-245', '2019-249', '2019-253', '2019-257', '2019-261', '2019-265', '2019-269', '2019-273', '2019-277', '2019-281', '2019-285', '2019-289', '2019-293', '2019-297', '2019-301', '2019-305', '2019-309', '2019-313', '2019-317', '2019-321', '2019-325', '2019-329', '2019-333', '2019-337', '2019-341', '2019-345', '2019-349', '2019-353', '2019-357', '2019-361', '2019-365']
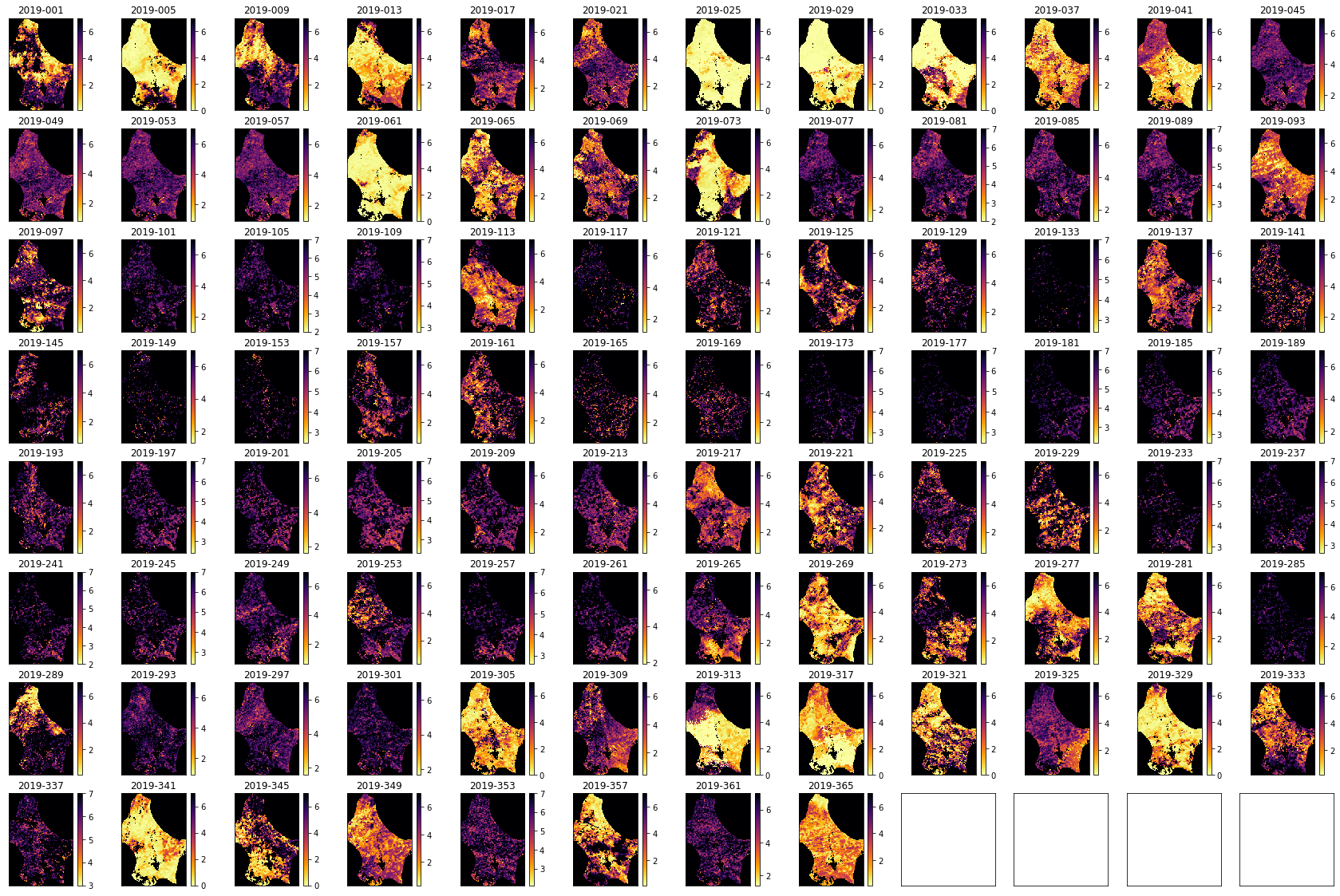
import matplotlib.pyplot as plt
#ANSWER
shape=(8,12)
x_size,y_size=(30,20)
fig, axs = plt.subplots(*shape,figsize=(x_size,y_size))
axs = axs.flatten()
plt.setp(axs, xticks=[], yticks=[])
for i in range(data.shape[0]):
im = axs[i].imshow(data[i],vmax=7,cmap=plt.cm.inferno_r,\
interpolation='nearest')
axs[i].set_title(bnames[i])
fig.colorbar(im, ax=axs[i])
Exercise 3
- Write a function
getLaithat takes as argument:year : integer year tile : list of tiles to process fips : country fips code (e.g. BE for Belgium)
and returns the annual LAI, standard deviation and day of year
- test your code for Belgium for 2018 for tiles
['h17v03','h18v03','h17v04','h18v04'] - show the shape of the arrays returned
Hint: You may find it useful to use modisAnnual
# ANSWER
import numpy as np
from geog0111.modisUtils import modisAnnual
from osgeo import gdal
def getLai(year=2019,tile=['h18v03','h18v04'],country='LU',verbose=False):
'''
Get LAI and std for year,tile,country
Options:
You should fill these out!!
'''
warp_args = {
'dstNodata' : 255,
'format' : 'MEM',
'cropToCutline' : True,
'cutlineWhere' : f"FIPS='{country}'",
'cutlineDSName' : 'data/TM_WORLD_BORDERS-0.3.shp'
}
kwargs = {
'tile' : tile,
'product' : 'MCD15A3H',
'sds' : ['Lai_500m','LaiStdDev_500m']
,
'doys' : np.arange(1,366,4),
'year' : year,
'warp_args' : warp_args,
'verbose' : False
}
# run
if verbose:
print(f'gathering modis annual data for {kwargs}')
odict,bnames = modisAnnual(**kwargs)
# read the data
if verbose:
print(f'reading datasets')
ddict = {}
for k,v in odict.items():
if verbose:
print(f'...{k} -> {v}')
g = gdal.Open(v)
if g:
ddict[k] = g.ReadAsArray()
# scale it
lai = ddict['Lai_500m'] * 0.1
std = ddict['LaiStdDev_500m'] * 0.1
# doy from filenames
doy = np.array([int(i.split('-')[1]) for i in bnames])
if verbose:
print(f'done')
return lai,std,doy
# Test
tile = ['h17v03','h18v03','h17v04','h18v04']
year = 2018
fips = 'BE'
# test your code for Belgium for 2018 for
# tiles ['h17v03','h18v03','h17v04','h18v04']
lai,std,doy = getLai(year,tile,fips,verbose=True)
print(f'shape of lai: {lai.shape}')
print(f'shape of std: {std.shape}')
print(f'shape of doy: {doy.shape}')
gathering modis annual data for {'tile': ['h17v03', 'h18v03', 'h17v04', 'h18v04'], 'product': 'MCD15A3H', 'sds': ['Lai_500m', 'LaiStdDev_500m'], 'doys': array([ 1, 5, 9, 13, 17, 21, 25, 29, 33, 37, 41, 45, 49,
53, 57, 61, 65, 69, 73, 77, 81, 85, 89, 93, 97, 101,
105, 109, 113, 117, 121, 125, 129, 133, 137, 141, 145, 149, 153,
157, 161, 165, 169, 173, 177, 181, 185, 189, 193, 197, 201, 205,
209, 213, 217, 221, 225, 229, 233, 237, 241, 245, 249, 253, 257,
261, 265, 269, 273, 277, 281, 285, 289, 293, 297, 301, 305, 309,
313, 317, 321, 325, 329, 333, 337, 341, 345, 349, 353, 357, 361,
365]), 'year': 2018, 'warp_args': {'dstNodata': 255, 'format': 'MEM', 'cropToCutline': True, 'cutlineWhere': "FIPS='BE'", 'cutlineDSName': 'data/TM_WORLD_BORDERS-0.3.shp'}, 'verbose': False}
reading datasets
...Lai_500m -> work/output_filename_Selektor_FIPS_BE_YEAR_2018_DOYS_1_365_SDS_Lai_500m.vrt
...LaiStdDev_500m -> work/output_filename_Selektor_FIPS_BE_YEAR_2018_DOYS_1_365_SDS_LaiStdDev_500m.vrt
Last update:
December 6, 2022